The “Collaborative Mini-Grids for Prediction of Viral RNA Structure and Evolution” project (in short: Mini-Grid project) aimed at designing a voluntary, peer-to-peer software architecture for executing parallelized bioinformatics algorithms, which makes research into RNA-based diseases like HIV, SARS, and bird flu more efficient than with current approaches. The project also studied how users can become aware of such an infrastructure, and designs the future technologies for biologist to use in the lab.
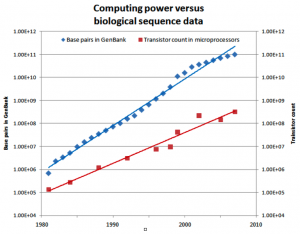
Biological sequence analysis suffers from a fundamental problem, namely that the amount of biological data available is growing faster than the computational power given by Moore’s Law (see Figure 1). This means that new, innovative methods must be developed that exploit the resources available for extensive calculations – for example grid computing.
In this project, the objective was to support biological research on viral RNA structure by utilizing local computing resources for distributed and collaborative bioinformatics computation and database search. More specifically, the project consisted of the following sub-projects:
- Creating a distributed and robust peer-to-peer software architecture for collaborative, distributed computation running on ordinary personal computers. This architecture was designed to be a general-purpose distribution platform for bioinformatics algorithms, which is used in this specific project to perform large-scale RNA structure prediction.
- Redesign and implement existing RNA structure prediction algorithms to make them more suitable for parallelization and distribution, and use the enhanced calculation capacity to predict the global RNA structures of virus genomes of HIV, SARS and bird flu.
- Experimental verification of the RNA structure predictions in the molecular biology laboratory. Novel RNA structures were synthesized and investigated by biochemical structure probing and analyzed at the single molecule level by Atomic Force Microscopy.
- Designing novel user interface technology for using the new grid technology and supporting the use of bioinformatics software in daily biology work.
Technology
I was personally involved in sub-project #1 and #4, i.e. in the design of the mini-grid technology and the novel user interface technology for lab work. The technological contributions included the following:
- The collaborative mini-grid infrastructure – a peer-to-peer grid infrastructure for distributed and parallel bioinformatics computation on office desktop and laptop computers [1, 2, 3].
- The GridOrbit awareness display – an interactive wall-based display providing an awareness of a grid infrastructure helping to recruit volunteer to the mini-grid [4, 5, 6].
- The Electronic Laboratory Workbench (eLabBench) – an interactive laboratory workbench integrating wet-lab biology experimentation with digital bioinformatics analysis [7, 8, 9].
- The so-called RABBIT technology for integrating biology lab test tubes with the eLabBench [10].
The eLabBench and the RABBIT is shown in the video below (in Danish).
Partners
The partners involved the IT University of Copenhagen (ITU), the Department of Molecular Biology and the interdisciplinary nanoscience centre (iNANO) at the University of Aarhus (AU), and CLC Bio A/S.
Funding & Time Period
 This research was funded byt the The Danish Council for Strategic Research from 2011–2016.
This research was funded byt the The Danish Council for Strategic Research from 2011–2016.
References
![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@Inbook{gpc2010:bardram,
author="Bardram, Jakob E.
and Venkataraman, Neelanarayanan",
editor="Bellavista, Paolo
and Chang, Ruay-Shiung
and Chao, Han-Chieh
and Lin, Shin-Feng
and Sloot, Peter M. A.",
title="The Mini-Grid Framework: Application Programming Support for Ad-Hoc, Peer-to-Peer Volunteer Grids",
bookTitle="Advances in Grid and Pervasive Computing: 5th International Conference, GPC 2010, Hualien, Taiwan, May 10-13, 2010. Proceedings",
year="2010",
publisher="Springer Berlin Heidelberg",
address="Berlin, Heidelberg",
pages="69--80",
abstract="Biological scientists work with bioinformatics algorithms that are either computational or data intensive in nature. Distributed platforms such as Grids and Peer-to-Peer (P2P) networks can be used for such algorithms. Classical Grid Computing platforms are available only to a restricted group of biologist, since they are expensive, and require skilled professionals for deployment and maintenance. Projects deployed using volunteer computing systems require a high visibility. The alternative, peer-to-peer platform is mainly used for sharing data. This paper presents the Mini-Grid Framework, a P2P infrastructure and programming framework for distribution of computational tasks like bioinformatics algorithm. The framework contributes with concepts and technologies for minimal configuration by non-technical end-users, a `resource-push' auction approach for dynamic task distribution, and context modeling of tasks and resources in order to handle volatile execution environment. The efficiency of the infrastructure has been evaluated using experiments.",
isbn="978-3-642-13067-0",
doi="10.1007/978-3-642-13067-0_11",
url="https://doi.org/10.1007/978-3-642-13067-0_11"
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@article{pcs2015:neela,
title = "A Context-aware Task Scheduling in Ad-hoc Desktop Grids",
journal = "Procedia Computer Science",
volume = "50",
number = "Supplement C",
pages = "653 - 662",
year = "2015",
note = "Big Data, Cloud and Computing Challenges",
issn = "1877-0509",
doi = "10.1016/j.procs.2015.04.099",
url = "https://doi.org/10.1016/j.procs.2015.04.099",
author = "Neelanarayanan Venkataraman",
keywords = "context information management, desktop grid computing, resource description, quality of service"
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png) N. N. Venkataraman, “The Mini-Grid Framework: Application Programming Support for Ad hoc Volunteer Grids,” PhD Thesis, 2013.
N. N. Venkataraman, “The Mini-Grid Framework: Application Programming Support for Ad hoc Volunteer Grids,” PhD Thesis, 2013. [Bibtex]
@phdthesis{phdthesis:neela,
title = "The Mini-Grid Framework: Application Programming Support for Ad hoc Volunteer Grids",
abstract = "To harvest idle, unused computational resources in networked environments, researchers have proposed different architectures for desktop grid infrastructure. However, most of the existing research work focus on centralized approach. In this thesis, we present the development and deployment of one such infrastructure, called the Mini-Grid Framework for resource management in ad hoc grids using market-based scheduling and context-based resource and application modeling. The framework proposes peer-to-peer architecture that supports several futures: ease of deployment, decentralized task distribution, small scale ad hoc grid formation, and symmetric resource. Furthermore, users can model and specify non-performance based parameters that influence resource allocation. We evaluated the framework through simulation experiments at the IT University of Copenhagen (ITU), as well as a pilot deployment at the Interdisciplinary Nanoscience Center (iNano), Aarhus University. For the simulation experiments we used an application that calculates prime numbers between a given range, and another application that searches for a key in a large data set. In the simulation experiments, we studied the technical performance and overhead of the Mini-Grid Framework and compared its performance with other relevant systems. For the pilot deployment, we have integrated a parallelized version of the Basic Local Alignment Search Tool (BLAST) algorithm and the RNA secondary structure prediction algorithm developed by iNano research center with the Mini-Grid Framework. These algorithms have been developed on top of our framework through an integration of the framework with the CLC Bio Workbench, a software suite for bioinformatics algorithms. The pilot deployment studied the resource participation, the deployment efforts needed, and the performance of the framework in a real grid environment. The main contribution of this thesis are: i) modeling entities such as resources and applications using their context, ii) the context-based auction strategy for dynamic task distribution, iii) scheduling through application specific quality parameters, iv) the definition of an extensible API for ad hoc grid formation and v) enabling symmetric resource participation. The Mini-Grid framework has been designed and developed as proof-of-concept. The Mini-Grid framework has been evaluated using LAB deployment at ITU, and has been deployed at iNano research center using real-life application.",
keywords = "Peer-to-peer Grids, Context-aware Scheduling, Context Modelling, Volunteer Grids, Application Specific Quality of Service",
author = "Venkataraman, Neelanarayanan Narayanan",
year = "2013",
isbn = "978-87-7949-292-9",
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p1899-hincapie-ramos,
author = {Hincapie-Ramos, Juan David and Tabard, Aurelien and Bardram, Jakob E.},
title = {GridOrbit: an infrastructure awareness system for increasing contribution in volunteer computing},
booktitle = {Proceedings of the 2011 annual conference on Human factors in computing systems},
series = {CHI '11},
year = {2011},
isbn = {978-1-4503-0228-9},
location = {Vancouver, BC, Canada},
pages = {1899--1908},
numpages = {10},
url = {http://doi.acm.org/10.1145/1978942.1979218},
doi = {10.1145/1978942.1979218},
acmid = {1979218},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {ambient displays, infrastructure awareness, infrastructures, public displays, volunteer computing},
tag={mini-grid,conference},
pdf={p1899-hincapie-ramos.pdf},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p3265-hincapie-ramos,
author = {Hincapie-Ramos, Juan David and Tabard, Aurelien and Bardram, Jakob E. and Sokoler, Tomas},
title = {GridOrbit public display: providing grid awareness in a biology laboratory},
booktitle = {CHI Extended Abstracts 2010},
year = {2010},
isbn = {978-1-60558-930-5},
pages = {3265--3270},
location = {Atlanta, Georgia, USA},
doi = {10.1145/1753846.1753969},
url = {http://doi.acm.org/10.1145/1753846.1753969},
publisher = {ACM},
address = {New York, NY, USA},
tag = {mini-grid,conference},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p302-hincapie-ramos,
author = {Hincapie-Ramos, Juan David and Tabard, Aurelien and Bardram, Jakob E.},
title = {Designing for the invisible: user-centered design of infrastructure awareness systems},
booktitle = {Proceedings of the 8th ACM Conference on Designing Interactive Systems},
series = {DIS '10},
year = {2010},
isbn = {978-1-4503-0103-9},
location = {Aarhus, Denmark},
pages = {302--305},
numpages = {4},
url = {http://doi.acm.org/10.1145/1858171.1858225},
doi = {10.1145/1858171.1858225},
acmid = {1858225},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {AMCards, infrastructure awareness, user-centred design},
tag = {mini-grid,conference},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{its2011:tabard,
author = {Tabard, Aur{\'e}lien and Hincapi{\'e}-Ramos, Juan-David and Esbensen, Morten and Bardram, Jakob E.},
title = {The eLabBench: An Interactive Tabletop System for the Biology Laboratory},
booktitle = {Proceedings of the ACM International Conference on Interactive Tabletops and Surfaces},
series = {ITS '11},
year = {2011},
isbn = {978-1-4503-0871-7},
location = {Kobe, Japan},
pages = {202--211},
numpages = {10},
url = {http://doi.acm.org/10.1145/2076354.2076391},
doi = {10.1145/2076354.2076391},
acmid = {2076391},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {activity-based computing, bench, biology, digital notebook, laboratory, tabletop},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p3051-tabard,
author = {Tabard, Aurelien and Hincapie Ramos, Juan David and Bardram, Jakob E},
title = {The eLabBench in the wild: supporting exploration in a molecular biology lab},
booktitle = {Proceedings of the 2012 ACM annual conference on Human Factors in Computing Systems},
series = {CHI '12},
year = {2012},
isbn = {978-1-4503-1015-4},
location = {Austin, Texas, USA},
pages = {3051--3060},
numpages = {10},
url = {http://doi.acm.org/10.1145/2207676.2208718},
doi = {10.1145/2207676.2208718},
acmid = {2208718},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {biology, design, digital bench, elabbench, experiment, field study, laboratory, life-sciences, research practices, tabletop},
tag={mini-grid,elabbench,conference},
pdf={p3051-tabard.pdf},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p251-tabard,
author = {Tabard, Aurelien and Gurn, Simon and Butz, Andreas and Bardram, Jakob E.},
title = {A case study of object and occlusion management on the eLabBench, a mixed physical/digital tabletop},
booktitle = {Proceedings of the 2013 ACM international conference on Interactive tabletops and surfaces},
series = {ITS '13},
year = {2013},
isbn = {978-1-4503-2271-3},
location = {St. Andrews, Scotland, United Kingdom},
pages = {251--254},
numpages = {4},
url = {http://doi.acm.org/10.1145/2512349.2512794},
doi = {10.1145/2512349.2512794},
acmid = {2512794},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {laboratory bench, object management, occlusion, tabletop, tangible interaction},
tag={mini-grid,elabbench,conference},
pdf={p251-tabard.pdf},
}![[pdf]](https://www.bardram.net/wp-content/plugins/papercite/img/pdf.png)
[Bibtex]
@inproceedings{p301-ramos,
author = {Hincapie-Ramos, Juan David and Tabard, Aurelien and Bardram, Jakob E.},
title = {Mediated tabletop interaction in the biology lab: exploring the design space of the rabbit},
booktitle = {Proceedings of the 13th international conference on Ubiquitous computing},
series = {UbiComp '11},
year = {2011},
isbn = {978-1-4503-0630-0},
location = {Beijing, China},
pages = {301--310},
numpages = {10},
url = {http://doi.acm.org/10.1145/2030112.2030153},
doi = {10.1145/2030112.2030153},
acmid = {2030153},
publisher = {ACM},
address = {New York, NY, USA},
keywords = {interactive surfaces, mediated tabletop interaction, rfid, tabletop, tangible, the rabbit},
tag={mini-grid,conference},
pdf={p301-ramos.pdf},
}
